Explaining Object classification using SHAP#
In this notebook, we will segment objects in an image and classify them using shape measurements. Using SHAP Analysis we can explain what role the different shape descriptors play.
import numpy as np
from skimage.io import imread
from skimage.measure import label, regionprops_table
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
import shap
import stackview
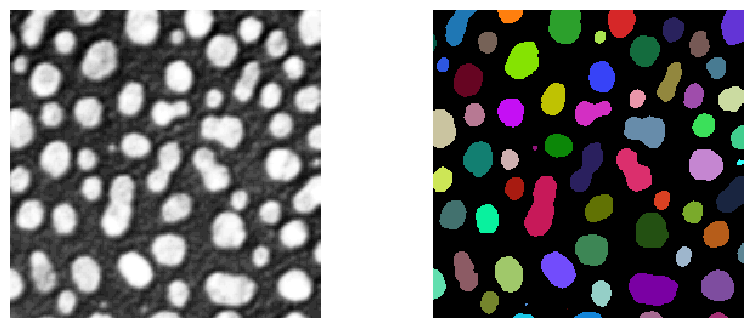
We use the blobs example image which contains multiple objects which could be grouped according to their size and shape. For example the 8-shaped objects in the center could be identified as group.
# Load the image
image = imread('data/blobs.tif')
# Apply threshold and
binary = image > 120
# Label connected components
labels = label(binary)
import stackview
from matplotlib import pyplot as plt
fig, ax = plt.subplots(1, 2, figsize=(10, 4))
stackview.imshow(image, plot=ax[0])
stackview.imshow(labels, plot=ax[1])
Extract region properties#
To allow object classification using shape features, we need to measure and compute the features.
# Measure object properties using scikit-image
properties = ['label', 'area', 'perimeter', 'mean_intensity', 'max_intensity',
'min_intensity', 'eccentricity', 'solidity', 'extent', "minor_axis_length", "major_axis_length"]
measurements = regionprops_table(labels, intensity_image=image, properties=properties)
df = pd.DataFrame(measurements)
# Add aspect ratio column
df['aspect_ratio'] = df['major_axis_length'] / df['minor_axis_length']
display(df.head())
| label | area | perimeter | mean_intensity | max_intensity | min_intensity | eccentricity | solidity | extent | minor_axis_length | major_axis_length | aspect_ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 433.0 | 91.254834 | 190.854503 | 232.0 | 128.0 | 0.876649 | 0.881874 | 0.555128 | 16.819060 | 34.957399 | 2.078439 |
| 1 | 2 | 185.0 | 53.556349 | 179.286486 | 224.0 | 128.0 | 0.828189 | 0.968586 | 0.800866 | 11.803854 | 21.061417 | 1.784283 |
| 2 | 3 | 658.0 | 95.698485 | 205.617021 | 248.0 | 128.0 | 0.352060 | 0.977712 | 0.870370 | 28.278264 | 30.212552 | 1.068402 |
| 3 | 4 | 434.0 | 76.870058 | 217.327189 | 248.0 | 128.0 | 0.341084 | 0.973094 | 0.820416 | 23.064079 | 24.535398 | 1.063793 |
| 4 | 5 | 477.0 | 83.798990 | 212.142558 | 248.0 | 128.0 | 0.771328 | 0.977459 | 0.865699 | 19.833058 | 31.162612 | 1.571246 |
Annotation data#
Next we load some annotation data. The annotation was hand-drawn on a label image and needs to be converted to tabular format first.
# Load annotation image and extract maximum intensity per label
annotation_image = imread('data/blobs_label_annotation.tif')
annotation_props = regionprops_table(labels, intensity_image=annotation_image,
properties=['label', 'max_intensity'])
annotation_df = pd.DataFrame(annotation_props)
annotation_df = annotation_df.rename(columns={'max_intensity': 'annotation'})
# Merge with main dataframe
df = df.merge(annotation_df, on='label')
display(df.head())
| label | area | perimeter | mean_intensity | max_intensity | min_intensity | eccentricity | solidity | extent | minor_axis_length | major_axis_length | aspect_ratio | annotation | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 433.0 | 91.254834 | 190.854503 | 232.0 | 128.0 | 0.876649 | 0.881874 | 0.555128 | 16.819060 | 34.957399 | 2.078439 | 0.0 |
| 1 | 2 | 185.0 | 53.556349 | 179.286486 | 224.0 | 128.0 | 0.828189 | 0.968586 | 0.800866 | 11.803854 | 21.061417 | 1.784283 | 0.0 |
| 2 | 3 | 658.0 | 95.698485 | 205.617021 | 248.0 | 128.0 | 0.352060 | 0.977712 | 0.870370 | 28.278264 | 30.212552 | 1.068402 | 2.0 |
| 3 | 4 | 434.0 | 76.870058 | 217.327189 | 248.0 | 128.0 | 0.341084 | 0.973094 | 0.820416 | 23.064079 | 24.535398 | 1.063793 | 0.0 |
| 4 | 5 | 477.0 | 83.798990 | 212.142558 | 248.0 | 128.0 | 0.771328 | 0.977459 | 0.865699 | 19.833058 | 31.162612 | 1.571246 | 0.0 |
len(df)
64
Train Random Forest Classifier#
Next, we train a random forest classifier. Therefore, we exctract only the objects which were annotated.
annotated_df = df[df['annotation'] != 0]
len(annotated_df)
12
# Prepare data for classification
feature_columns = ["solidity", "perimeter", "area", "aspect_ratio", "extent"]
# annotated_df.columns #['area', 'perimeter', 'mean_intensity', 'max_intensity',
# 'min_intensity', 'eccentricity', 'solidity', 'extent']
X = annotated_df[feature_columns]
y = annotated_df['annotation']
# Split data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Train model
rf = RandomForestClassifier(n_estimators=100, random_state=42)
rf.fit(X_train, y_train)
# Print accuracy
print(f"Training accuracy: {rf.score(X_train, y_train):.3f}")
print(f"Testing accuracy: {rf.score(X_test, y_test):.3f}")
Training accuracy: 1.000
Testing accuracy: 0.333
We can now apply this classifier to the entire dataset.
y_ = rf.predict(df[feature_columns])
y_
array([1., 3., 3., 3., 3., 3., 2., 3., 3., 1., 3., 3., 2., 3., 3., 1., 3.,
3., 3., 3., 3., 3., 1., 3., 3., 3., 1., 3., 3., 3., 1., 2., 1., 3.,
3., 1., 3., 1., 3., 3., 2., 3., 3., 3., 3., 3., 3., 3., 1., 3., 3.,
1., 3., 3., 3., 1., 3., 3., 3., 2., 1., 3., 1., 1.])
# Map labels to y values
result = labels.copy()
for i, label_id in enumerate(np.unique(labels)[1:], 1): # skip 0 as it's background
result[labels == label_id] = y_[i-1]
# Show result
stackview.insight(result)
|
|
|
Explain classification using SHAP values#
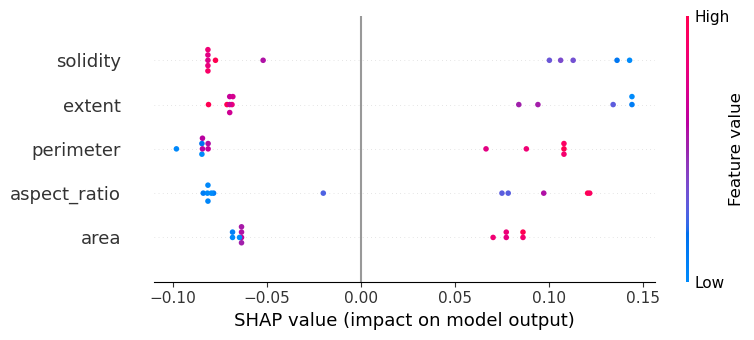
Using the SHAP-plot we can determine which features contribute most to the decision of the classifier. The plot below can be interpreted like this:
The
solidityandextendfeatures contribute most to the classification. If solidity abd extend are low (blue), the object might be 8-shaped.Also
perimeterandaspect_ratiocontribute. If they are high, the object might be 8-shaped.The
areacontributes as well, just a little les prominently. If objects are large, they are more likely to be 8-shaped.
explainer = shap.TreeExplainer(rf)
shap_values = explainer.shap_values(X)[...,0]
shap.summary_plot(shap_values, X) #, feature_names=feature_columns)
Exercise#
Draw the SHAP summary plot for the shap values [..., 1]. Which object class was this SHAP plot drawn for?
