Simulation of image formation + image restoration#
In this notebook we artificially assemble a microscope image from simulated nuclei, noise and background. Afterwards, we use classical image processing techniques to remove noise and background.
import pyclesperanto_prototype as cle
import numpy as np
image_size = (100, 100)
# noise configuration
noise_level = 2
# background configuration
camera_offset = 100
background_sigma = 25
background_intensity = 5
# nuclei configuration
nuclei_radius = 5
nuclei_blur_sigma = 1
nuclei_number = 10
nuclei_intensity = 5
# by pinning the random seed, we can make the code repeatable
np.random.seed(42)
Noise#
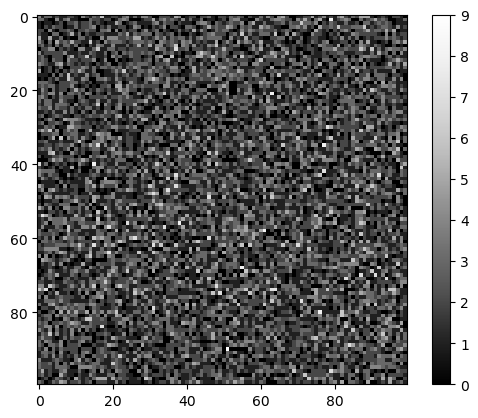
Here we assume that the noise in the image is Poisson distributed, a common assumtion in microscopy.
noise_image = np.random.poisson(noise_level, image_size)
cle.imshow(noise_image, colorbar=True)
Background#
Background intensity in fluorescence microscopy images typically comes from out-of-focus light. We can simulate this by placing light sources as single pixels and blurring them with a Gaussian filter. Furthermore, many microscope cameras have a so called camera offset. No pixel will ever have intensity below this value.
# create empty image
background = np.zeros(image_size)
# place light sources
background[20, 10] += 1
background[50, 80] += 1
background[60, 50] += 1
# blur them massively
background = cle.gaussian_blur(background, sigma_x=background_sigma, sigma_y=background_sigma)
# normalize the image so that the maximum intensity has a defined value
background = background / background.max() * background_intensity
# add camera offsert
background = background + camera_offset
background
|
|
cle._ image
|
Nuclei#
Next we place nuclei in an image at random positions. We blur them a bit to simulate the point-spread-function of the microscope.
# retrieve a defined number of random positions
nuclei_positions = np.random.random((nuclei_number, 2)) * image_size
# write 1 at these locations
nuclei_image = cle.pointlist_to_labelled_spots(nuclei_positions.T, np.zeros(image_size))
nuclei_image = (nuclei_image > 0) * nuclei_intensity
# enlarge the nuclei by a define radius
nuclei_image = cle.maximum_sphere(nuclei_image, radius_x=nuclei_radius, radius_y=nuclei_radius)
# blur the image to make it look more realistic
nuclei_image = cle.gaussian_blur(nuclei_image, sigma_x=nuclei_blur_sigma, sigma_y=nuclei_blur_sigma)
nuclei_image
|
|
cle._ image
|
Image formation#
A microscopy image is the sum of the scence and the effect described above.
sum_image = np.asarray(noise_image + background + nuclei_image)
cle.imshow(sum_image, colorbar=True)
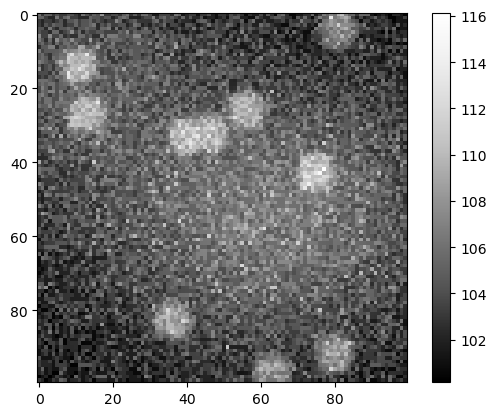
Image segmentation#
If we now applied a segmentation algorithm to this image as it is, it might lead to a wrong result.
binary = cle.threshold_otsu(sum_image.astype(np.float32))
binary
|
|
cle._ image
|
Background removal#
To fix this problem, we need to remove the background intensity first.
background_removed = cle.top_hat_box(sum_image, radius_x=10, radius_y=10)
background_removed
|
|
cle._ image
|
Noise removal#
We can also remove the noise from the image.
noise_removed1 = cle.mean_sphere(sum_image, radius_x=3, radius_y=3)
noise_removed1
|
|
cle._ image
|
And this can also be done on the background-subtracted image.
noise_removed = cle.mean_sphere(background_removed, radius_x=3, radius_y=3)
noise_removed
|
|
cle._ image
|
Image segmentation II#
After correcting the image, we can try segmentation again.
binary2 = cle.threshold_otsu(noise_removed.astype(np.float32))
binary2
|
|
cle._ image
|
# sneak preview: watershed
import napari_segment_blobs_and_things_with_membranes as nsbatwm
binary3 = nsbatwm.split_touching_objects(binary2)
binary3
<__array_function__ internals>:180: RuntimeWarning: Converting input from bool to <class 'numpy.uint8'> for compatibility.
|
|
nsbatwm made image
|
