Comparison of measurements from different libraries#
We now compare the implementation of the measurement perimeter in napari-skimage-regionprops regionprops_table and napari-simpleitk-image-processing label_statistics.
import numpy as np
import pandas as pd
from skimage.io import imread
from pyclesperanto_prototype import imshow
from skimage import filters
from skimage import measure
import matplotlib.pyplot as plt
from napari_skimage_regionprops import regionprops_table
from skimage import filters
from napari_simpleitk_image_processing import label_statistics
C:\Users\maral\mambaforge\envs\feature_blogpost\lib\site-packages\morphometrics\measure\label.py:7: TqdmExperimentalWarning: Using `tqdm.autonotebook.tqdm` in notebook mode. Use `tqdm.tqdm` instead to force console mode (e.g. in jupyter console)
from tqdm.autonotebook import tqdm
Therefore, we need an image and a label image.
# load image
image = imread("../../data/blobs.tif")
# denoising
blurred_image = filters.gaussian(image, sigma=1)
# binarization
threshold = filters.threshold_otsu(blurred_image)
thresholded_image = blurred_image >= threshold
# labeling
label_image = measure.label(thresholded_image)
# visualization
imshow(label_image, labels=True)
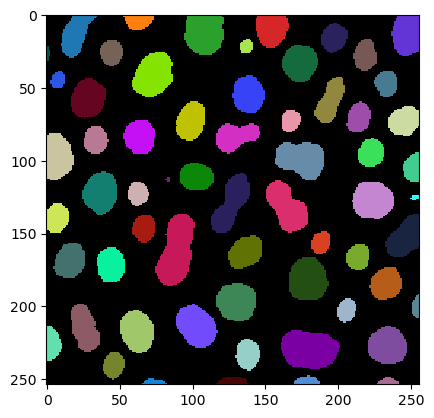
We derive the perimeter measurements using the regionprops_table function
skimage_statistics = regionprops_table(image, label_image, perimeter = True)
skimage_statistics
| label | area | bbox_area | equivalent_diameter | convex_area | max_intensity | mean_intensity | min_intensity | perimeter | perimeter_crofton | standard_deviation_intensity | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 429 | 750 | 23.371345 | 479 | 232.0 | 191.440559 | 128.0 | 89.012193 | 87.070368 | 29.793138 |
| 1 | 2 | 183 | 231 | 15.264430 | 190 | 224.0 | 179.846995 | 128.0 | 53.556349 | 53.456120 | 21.270534 |
| 2 | 3 | 658 | 756 | 28.944630 | 673 | 248.0 | 205.604863 | 120.0 | 95.698485 | 93.409370 | 29.392255 |
| 3 | 4 | 433 | 529 | 23.480049 | 445 | 248.0 | 217.515012 | 120.0 | 77.455844 | 76.114262 | 35.852345 |
| 4 | 5 | 472 | 551 | 24.514670 | 486 | 248.0 | 213.033898 | 128.0 | 83.798990 | 82.127941 | 28.741080 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 57 | 58 | 213 | 285 | 16.468152 | 221 | 224.0 | 184.525822 | 120.0 | 52.284271 | 52.250114 | 28.255467 |
| 58 | 59 | 79 | 108 | 10.029253 | 84 | 248.0 | 184.810127 | 128.0 | 39.313708 | 39.953250 | 33.739912 |
| 59 | 60 | 88 | 110 | 10.585135 | 92 | 216.0 | 182.727273 | 128.0 | 45.692388 | 46.196967 | 24.417173 |
| 60 | 61 | 52 | 75 | 8.136858 | 56 | 248.0 | 189.538462 | 128.0 | 30.692388 | 32.924135 | 37.867411 |
| 61 | 62 | 48 | 68 | 7.817640 | 53 | 224.0 | 173.833333 | 128.0 | 33.071068 | 35.375614 | 27.987596 |
62 rows × 11 columns
… and using the label_statistics function
simpleitk_statistics = label_statistics(image, label_image, size = False, intensity = False, perimeter = True)
simpleitk_statistics
| label | perimeter | perimeter_on_border | perimeter_on_border_ratio | |
|---|---|---|---|---|
| 0 | 1 | 87.070368 | 16.0 | 0.183759 |
| 1 | 2 | 53.456120 | 21.0 | 0.392846 |
| 2 | 3 | 93.409370 | 23.0 | 0.246228 |
| 3 | 4 | 76.114262 | 20.0 | 0.262763 |
| 4 | 5 | 82.127941 | 40.0 | 0.487045 |
| ... | ... | ... | ... | ... |
| 57 | 58 | 52.250114 | 0.0 | 0.000000 |
| 58 | 59 | 39.953250 | 18.0 | 0.450527 |
| 59 | 60 | 46.196967 | 22.0 | 0.476222 |
| 60 | 61 | 32.924135 | 15.0 | 0.455593 |
| 61 | 62 | 35.375614 | 17.0 | 0.480557 |
62 rows × 4 columns
For this comparison, we are only interested in the column named perimeter. So we are selecting this column and convert the included measurements into a numpy array:
skimage_perimeter = np.asarray(skimage_statistics['perimeter'])
simpleitk_perimeter = np.asarray(simpleitk_statistics['perimeter'])
Scatter plot#
If we now use matplotlib to plot the two perimeter measurements against each other, we receive the following scatter plot:
plt.plot(skimage_perimeter, simpleitk_perimeter, 'o')
[<matplotlib.lines.Line2D at 0x1912b551220>]
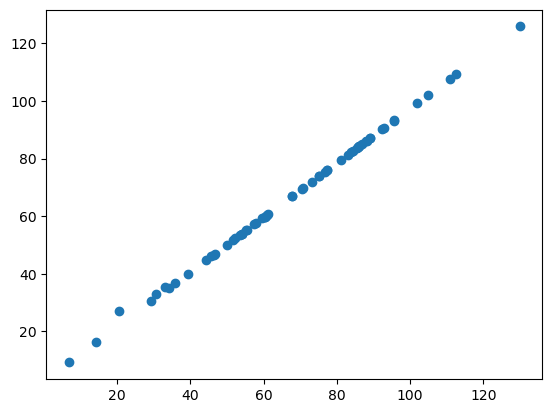
If the two libraries would compute the perimeter in the same way, all of our datapoints would lie on a straight line. As you can see, they are not. This suggests that perimeter is implemented differently in napari-skimage-regionprops and napari-simpleitk-image-processing.
Bland-Altman plot#
The Bland-Altman plot helps to visualize the difference between measurements. We can compute a Bland-Altman plot for our two different perimeter implementations like this:
# compute mean, diff, md and sd
mean = (skimage_perimeter + simpleitk_perimeter) / 2
diff = skimage_perimeter - simpleitk_perimeter
md = np.mean(diff) # mean of difference
sd = np.std(diff, axis = 0) # standard deviation of difference
# add mean and diff
plt.plot(mean, diff, 'o')
# add lines
plt.axhline(md, color='gray', linestyle='--')
plt.axhline(md + 1.96*sd, color='gray', linestyle='--')
plt.axhline(md - 1.96*sd, color='gray', linestyle='--')
# add title and axes labels
plt.title('Bland-Altman Plot')
plt.xlabel('Average Measurement')
plt.ylabel('Difference of Measurement')
Text(0, 0.5, 'Difference of Measurement')
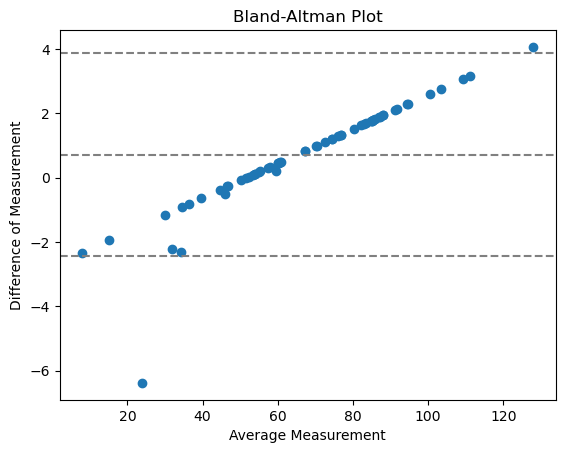
With
center line = mean difference between the methods
two outer lines = confidence interval of agreement (CI)
The points do not go towards 0 which means it is not an agreement with random relative error, but systematic. This makes a lot of sense, because we are comparing here the implementation of a measurement.
Exercise#
Use the functions regionprops_table and label_statistics to measure feret_diameter_max of your label image. Plot the Scatter plot and the Bland-Altman plot. Do you think feret_diameter_max is implemented differently in the two libraries?
