Terminology#
This notebook serves showing images and segmented images of different kind to explain common terminology.
from skimage.io import imread, imshow
import napari_segment_blobs_and_things_with_membranes as nsbatwm
import stackview
import numpy as np
Intensity images#
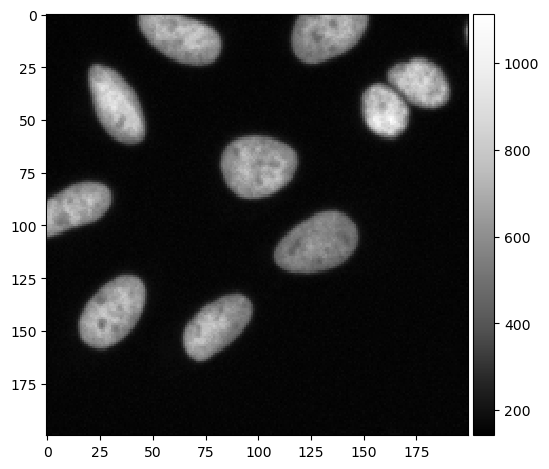
We start with intensity images as they are commonly produced by microscopes.
input_image = imread("../../data/BBBC022/IXMtest_A02_s9.tif")[:,:,0]
cropped_image = input_image[0:200, 200:400]
imshow(cropped_image, cmap='Greys_r')
C:\Users\haase\mambaforge\envs\bio39\lib\site-packages\skimage\io\_plugins\matplotlib_plugin.py:150: UserWarning: Low image data range; displaying image with stretched contrast.
lo, hi, cmap = _get_display_range(image)
<matplotlib.image.AxesImage at 0x2ba68647e20>
Binary images#
Basic segmentation algorithms lead to binary images. They are characterized by having only two different intensities such as 0 and 1.
binary_image = nsbatwm.threshold_otsu(cropped_image)
binary_image
|
|
nsbatwm made image
|
Instance segmentation#
If each individual object has its own intensity / color, we call it an instance segmentation label image. The inistances are labeled with integer numbers. The maximum intensity (label) in this image typically corresponds to the number of objects.
instance_segmentation = nsbatwm.voronoi_otsu_labeling(cropped_image,
spot_sigma=5,
outline_sigma=1)
instance_segmentation
|
|
nsbatwm made image
|
When you run this notebook locally, you can hover with your mouse over the image and inspect pixel intensities.
import stackview
stackview.picker(instance_segmentation)
Semantic segmentation#
Semantic segmentation label images can have more than two labels and typically label regions where pixels have the same meanings, for example: nuclei, nuclear envelope and background.
semantic_segmentation = binary_image + nsbatwm.maximum_filter(binary_image).astype(np.uint32) + 1
semantic_segmentation
|
|
nsbatwm made made image
|
Sparse annotations#
Annotated label images are typically drawn by humans. When executing this notebook locally, you can draw some annotations. If you annotate a couple of nuclei precisely, you create a sparse nuclei annotation. Hold down the ALT key to erase an annotation.
sparse_label_annotation = np.zeros_like(cropped_image, dtype=np.uint32)
stackview.annotate(cropped_image, sparse_label_annotation)
stackview.insight(sparse_label_annotation)
|
|
|
You can also create a sparse semantic annotation, e.g. by annotating some pixels within nuclei and some pixels within the background.
sparse_semantic_annotation = np.zeros_like(cropped_image, dtype=np.uint32)
stackview.annotate(cropped_image, sparse_semantic_annotation)
stackview.insight(sparse_semantic_annotation)
|
|
|
