Differentiating nuclei according to signal intensity#
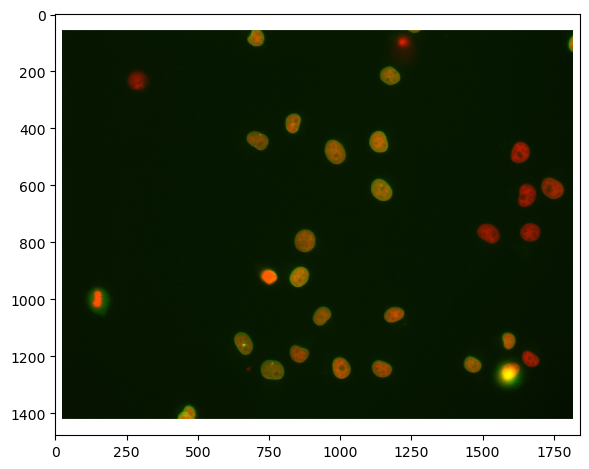
A common bio-image analysis task is differentiating cells according to their signal expression. In this example we take a two-channel image of nuclei which express Cy3 and eGFP. Visually, we can easily see that some nuclei expressing Cy3 also express eGFP, others don’t. This notebook demonstrates how to differentiate nuclei segmented in one channel according to the intensity in the other channel.
import pyclesperanto_prototype as cle
import numpy as np
from skimage.io import imread, imshow
import matplotlib.pyplot as plt
import pandas as pd
cle.get_device()
<Intel(R) Iris(R) Xe Graphics on Platform: Intel(R) OpenCL HD Graphics (1 refs)>
We’re using a dataset published by Heriche et al. licensed CC BY 4.0 available in the Image Data Resource.
# load file
raw_image = imread('../../data/plate1_1_013 [Well 5, Field 1 (Spot 5)].png')
# visualize
imshow(raw_image)
<matplotlib.image.AxesImage at 0x1dac72d7c40>
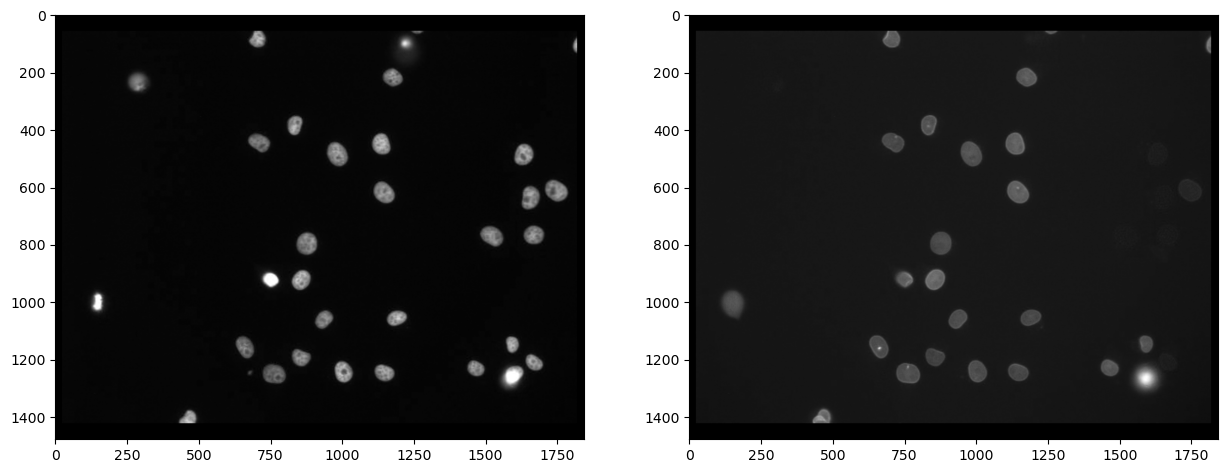
First, we need to split channels (read more). After that, we can actually see that not all cells marked with Cy3 (channel 0) are also marked with eGFP (channel 1):
# extract channels
channel_cy3 = raw_image[...,0]
channel_egfp = raw_image[...,1]
# visualize
fig, axs = plt.subplots(1, 2, figsize=(15,15))
axs[0].imshow(channel_cy3, cmap='gray')
axs[1].imshow(channel_egfp, cmap='gray')
<matplotlib.image.AxesImage at 0x1dac7328970>
Segmenting nuclei#
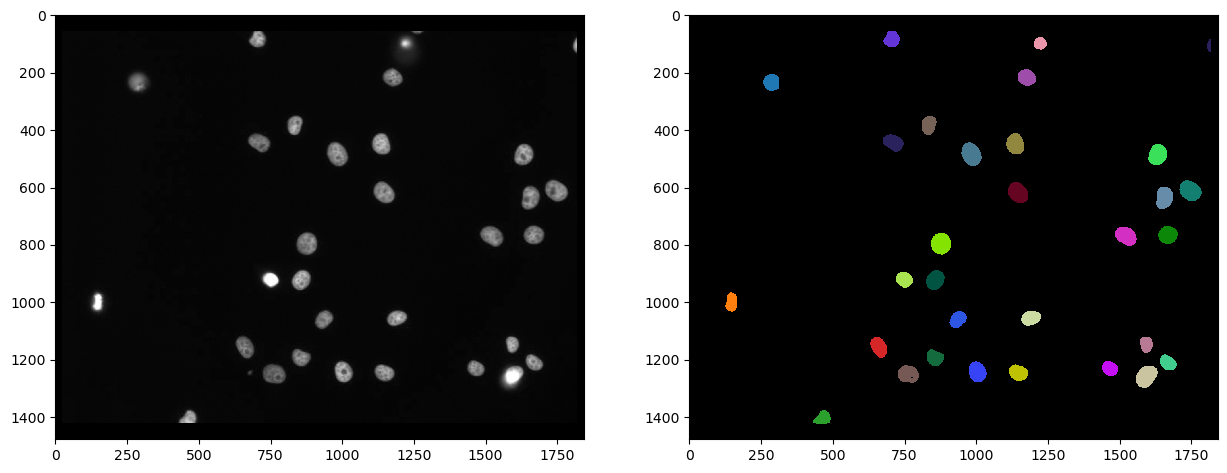
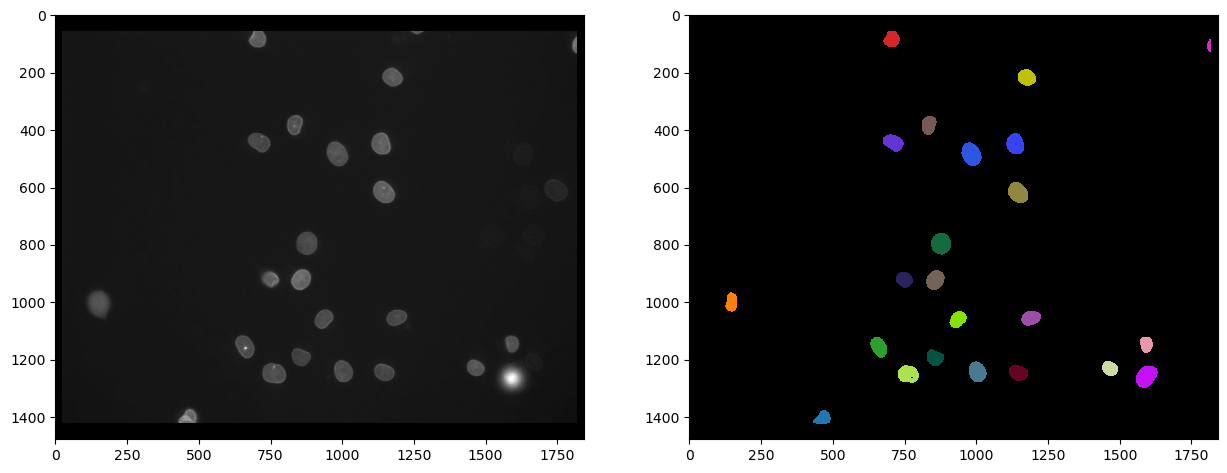
As the staining marks the nuclei in the Cy3 channel, it is reasonable to segment nuclei in this channel and afterwards measure the intensity in the other channel. We use Voronoi-Otsu-Labeling for the segmentation because it is a quick and straightforward approach.
# segmentation
nuclei_cy3 = cle.voronoi_otsu_labeling(channel_cy3, spot_sigma=20)
# visualize
fig, axs = plt.subplots(1, 2, figsize=(15,15))
cle.imshow(channel_cy3, plot=axs[0], color_map="gray")
cle.imshow(nuclei_cy3, plot=axs[1], labels=True)
C:\Users\rober\miniconda3\envs\bio39\lib\site-packages\pyclesperanto_prototype\_tier9\_imshow.py:46: UserWarning: The imshow parameter color_map is deprecated. Use colormap instead.
warnings.warn("The imshow parameter color_map is deprecated. Use colormap instead.")
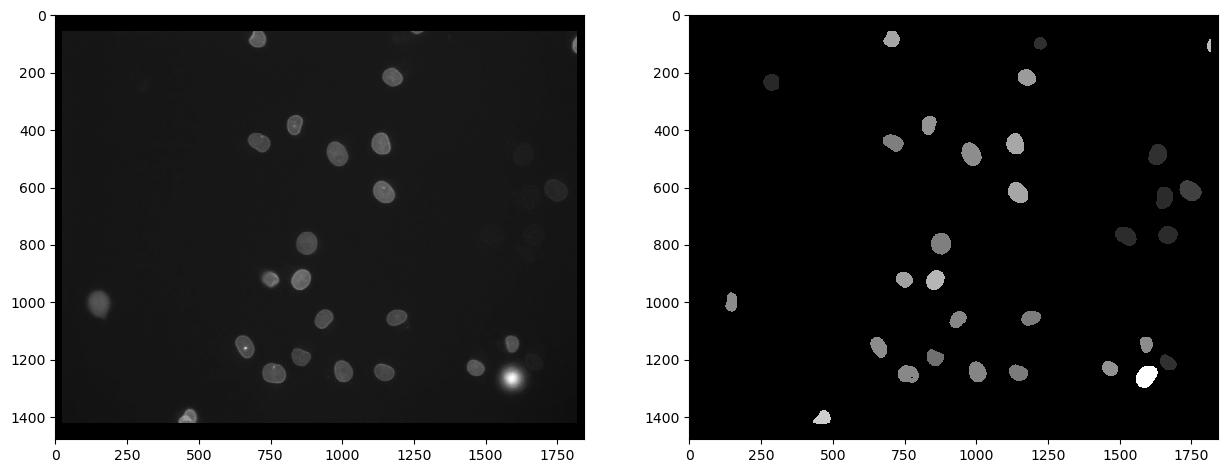
Firstly, we can measure the intensity in the second channel, marked with eGFP and visualize that measurement in a parametric image. In such a parametric image, all pixels inside a nucleus have the same value, in this case the mean average intensity of the cell.
intensity_map = cle.mean_intensity_map(channel_egfp, nuclei_cy3)
# visualize
fig, axs = plt.subplots(1, 2, figsize=(15,15))
cle.imshow(channel_egfp, plot=axs[0], color_map="gray")
cle.imshow(intensity_map, plot=axs[1], color_map="gray")
C:\Users\rober\miniconda3\envs\bio39\lib\site-packages\pyclesperanto_prototype\_tier9\_imshow.py:46: UserWarning: The imshow parameter color_map is deprecated. Use colormap instead.
warnings.warn("The imshow parameter color_map is deprecated. Use colormap instead.")
From such a parametric image, we can retrieve the intensity values and get them in a vector. The first item in this list has value 0 and corresponds to the intensity of the background, which is 0 in the parametric image.
intensity_vector = cle.read_intensities_from_map(nuclei_cy3, intensity_map)
intensity_vector
cle.array([[ 0. 80.875854 23.529799 118.17817 80.730255 95.55177 72.84752 92.34759 78.84362 85.400444 105.108025 65.06639 73.69295 77.40091 81.48371 77.12868 96.58209 95.94536 70.883995 89.70502 72.01046 27.257877 84.460075 25.49711 80.69057 147.49736 28.112642 25.167627 28.448263 25.31705 38.072815 108.81613 ]], dtype=float32)
There is by the way also an alternative way to come to the mean intensities directly, by measuring all properties of the nuclei including position and shape. The statistics can be further processed as pandas DataFrame.
statistics = cle.statistics_of_background_and_labelled_pixels(channel_egfp, nuclei_cy3)
statistics_df = pd.DataFrame(statistics)
statistics_df.head()
| label | original_label | bbox_min_x | bbox_min_y | bbox_min_z | bbox_max_x | bbox_max_y | bbox_max_z | bbox_width | bbox_height | ... | centroid_z | sum_distance_to_centroid | mean_distance_to_centroid | sum_distance_to_mass_center | mean_distance_to_mass_center | standard_deviation_intensity | max_distance_to_centroid | max_distance_to_mass_center | mean_max_distance_to_centroid_ratio | mean_max_distance_to_mass_center_ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 0.0 | 0.0 | 0.0 | 1841.0 | 1477.0 | 0.0 | 1842.0 | 1478.0 | ... | 0.0 | 1.683849e+09 | 640.648682 | 1.683884e+09 | 640.661682 | 8.487288 | 1187.033203 | 1187.743164 | 1.852861 | 1.853932 |
| 1 | 2 | 1 | 127.0 | 967.0 | 0.0 | 167.0 | 1033.0 | 0.0 | 41.0 | 67.0 | ... | 0.0 | 4.128044e+04 | 18.704323 | 4.128327e+04 | 18.705606 | 4.734930 | 34.280727 | 34.338104 | 1.832770 | 1.835712 |
| 2 | 3 | 2 | 259.0 | 205.0 | 0.0 | 314.0 | 265.0 | 0.0 | 56.0 | 61.0 | ... | 0.0 | 5.392080e+04 | 19.715099 | 5.393993e+04 | 19.722092 | 1.663603 | 32.079941 | 31.469477 | 1.627176 | 1.595646 |
| 3 | 4 | 3 | 432.0 | 1377.0 | 0.0 | 492.0 | 1423.0 | 0.0 | 61.0 | 47.0 | ... | 0.0 | 3.630314e+04 | 17.769527 | 3.636823e+04 | 17.801388 | 24.842560 | 36.856213 | 36.085457 | 2.074125 | 2.027115 |
| 4 | 5 | 4 | 631.0 | 1123.0 | 0.0 | 690.0 | 1194.0 | 0.0 | 60.0 | 72.0 | ... | 0.0 | 6.753254e+04 | 21.686750 | 6.755171e+04 | 21.692907 | 17.358543 | 38.805695 | 38.417568 | 1.789373 | 1.770974 |
5 rows × 37 columns
The intensity vector can then be retrieved from the tabular statistics. Note: In this case, the background intensity is not 0, because we were directly reading it from the original image.
intensity_vector2 = statistics['mean_intensity']
intensity_vector2
array([ 20.829758, 80.875854, 23.529799, 118.17817 , 80.730255,
95.55177 , 72.84752 , 92.34759 , 78.84362 , 85.400444,
105.108025, 65.06639 , 73.69295 , 77.40091 , 81.48371 ,
77.12868 , 96.58209 , 95.94536 , 70.883995, 89.70502 ,
72.01046 , 27.257877, 84.460075, 25.49711 , 80.69057 ,
147.49736 , 28.112642, 25.167627, 28.448263, 25.31705 ,
38.072815, 108.81613 ], dtype=float32)
To get an overview about the mean intensity measurement, we can use matplotlib to plot a histogram. We ignore the first element, because it corresponds to the background intensity.
plt.hist(intensity_vector2[1:])
(array([ 7., 1., 0., 4., 10., 5., 2., 1., 0., 1.]),
array([ 23.52979851, 35.92655563, 48.32331085, 60.72006607,
73.11682129, 85.51358032, 97.91033936, 110.30709076,
122.70384979, 135.1006012 , 147.49736023]),
<BarContainer object of 10 artists>)
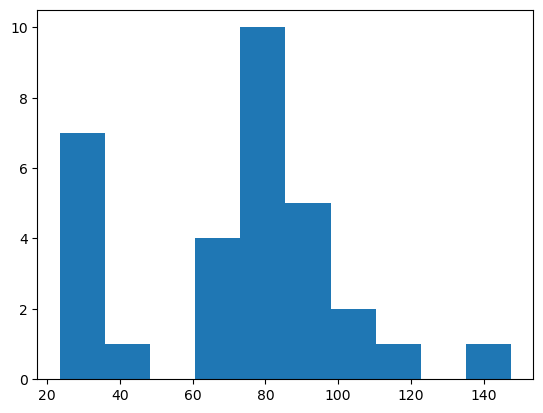
From such a histogram, we could conclude that objects with intensity above 50 are positive.
Selecting labels above a given intensity threshold#
We next generate a new labels image with the nuclei having intensity > 50. Note, all the above steps for extracting the intensity vector are not necessary for this. We just did that to get an idea about a good intensity threshold.
The following label image show the nuclei segmented in the Cy3 channel which have a high intensity in the eGFP channel.
intensity_threshold = 50
nuclei_with_high_intensity_egfg = cle.exclude_labels_with_map_values_within_range(intensity_map, nuclei_cy3, maximum_value_range=intensity_threshold)
intensity_map = cle.mean_intensity_map(channel_egfp, nuclei_cy3)
# visualize
fig, axs = plt.subplots(1, 2, figsize=(15,15))
cle.imshow(channel_egfp, plot=axs[0], color_map="gray")
cle.imshow(nuclei_with_high_intensity_egfg, plot=axs[1], labels=True)
C:\Users\rober\miniconda3\envs\bio39\lib\site-packages\pyclesperanto_prototype\_tier9\_imshow.py:46: UserWarning: The imshow parameter color_map is deprecated. Use colormap instead.
warnings.warn("The imshow parameter color_map is deprecated. Use colormap instead.")
And we can also count those by determining the maximum intensity in the label image:
number_of_double_positives = nuclei_with_high_intensity_egfg.max()
print("Number of Cy3 nuclei that also express eGFP", number_of_double_positives)
Number of Cy3 nuclei that also express eGFP 23.0
References#
Some of the functions we used might be uncommon. Hence, we can add their documentation for reference.
print(cle.voronoi_otsu_labeling.__doc__)
Labels objects directly from grey-value images.
The two sigma parameters allow tuning the segmentation result. Under the hood,
this filter applies two Gaussian blurs, spot detection, Otsu-thresholding [2] and Voronoi-labeling [3]. The
thresholded binary image is flooded using the Voronoi tesselation approach starting from the found local maxima.
Notes
-----
* This operation assumes input images are isotropic.
Parameters
----------
source : Image
Input grey-value image
label_image_destination : Image, optional
Output image
spot_sigma : float, optional
controls how close detected cells can be
outline_sigma : float, optional
controls how precise segmented objects are outlined.
Returns
-------
label_image_destination
Examples
--------
>>> import pyclesperanto_prototype as cle
>>> cle.voronoi_otsu_labeling(source, label_image_destination, 10, 2)
References
----------
.. [1] https://clij.github.io/clij2-docs/reference_voronoiOtsuLabeling
.. [2] https://ieeexplore.ieee.org/document/4310076
.. [3] https://en.wikipedia.org/wiki/Voronoi_diagram
print(cle.mean_intensity_map.__doc__)
Takes an image and a corresponding label map, determines the mean
intensity per label and replaces every label with the that number.
This results in a parametric image expressing mean object intensity.
Parameters
----------
source : Image
label_map : Image
destination : Image, optional
Returns
-------
destination
References
----------
.. [1] https://clij.github.io/clij2-docs/reference_meanIntensityMap
print(cle.read_intensities_from_map.__doc__)
Takes a label image and a parametric image to read parametric values from the labels positions.
The read intensity values are stored in a new vector.
Note: This will only work if all labels have number of voxels == 1 or if all pixels in each label have the same value.
Parameters
----------
labels: Image
map_image: Image
values_destination: Image, optional
1d vector with length == number of labels + 1
Returns
-------
values_destination, Image:
vector of intensity values with 0th element corresponding to background and subsequent entries corresponding to
the intensity in the given labeled object
print(cle.statistics_of_background_and_labelled_pixels.__doc__)
Determines bounding box, area (in pixels/voxels), min, max and mean
intensity of background and labelled objects in a label map and corresponding
pixels in the original image.
Instead of a label map, you can also use a binary image as a binary image is a
label map with just one label.
This method is executed on the CPU and not on the GPU/OpenCL device.
Parameters
----------
source : Image
labelmap : Image
Returns
-------
Dictionary of measurements
References
----------
.. [1] https://clij.github.io/clij2-docs/reference_statisticsOfBackgroundAndLabelledPixels
print(cle.exclude_labels_with_map_values_within_range.__doc__)
This operation removes labels from a labelmap and renumbers the
remaining labels.
Notes
-----
* Values of all pixels in a label each must be identical.
Parameters
----------
values_map : Image
label_map_input : Image
label_map_destination : Image, optional
minimum_value_range : Number, optional
maximum_value_range : Number, optional
Returns
-------
label_map_destination
References
----------
.. [1] https://clij.github.io/clij2-docs/reference_excludeLabelsWithValuesWithinRange
