Image Segmentation with CellPose#
CellPose is a deep-learning based segmentation algorithm for cells and nuclei in microscopy images.
See also
As usual, we start with loading an example image.
import stackview
from cellpose import models, io
import numpy as np
from skimage.data import human_mitosis
image = human_mitosis()
stackview.insight(image)
|
|
|
Loading a pretrained model#
CellPose comes with a number of pretrained models, e.g. for segmenting images showing cells or nuclei. We will just load a model for segmenting nuclei.
model = models.Cellpose(gpu=False, model_type='nuclei')
We let the model “evaluate” the image to produce masks of segmented nuclei.
channels = [0,0] # This means we are processing single-channel greyscale images.
masks, flows, styles, diams = model.eval(image, diameter=None, channels=channels)
stackview.insight(masks.astype(np.uint32))
|
|
|
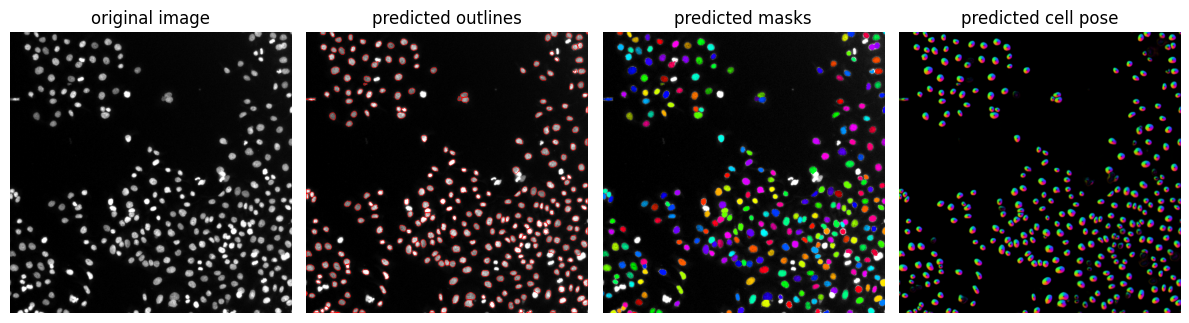
Result visualization#
Cell / nuclei segmentation results can be checked best if the resulting label image is overlaid to the original image or by drawing outlines around segmented regions.
from cellpose import plot
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(12,5))
plot.show_segmentation(fig, image, masks, flows[0], channels=channels)
plt.tight_layout()
plt.show()
Exercise#
Load ../../data/blobs.tif and apply Cellpose to it.
